Abstract
This article focuses on ECG signal recognition based on acoustic feature extraction techniques. The SVM and k-NN classification approaches are proposed for recognizing the ECG heart sound as well as for calculating the recognition efficiency. In this proposed technique, ECG signals are previously transformed into a successive series of Mel-frequency cepstral coefficients for computing the acoustic features in terms of mean value. A histogram based understandable and new approach is proposed at this point for recognition of ‘P’ wave, ‘R’ wave etc. from ECG waveform. The recognition of ECG signal and their distinguishing features provide significant effort for the analysis. Here three statistical data with their detection efficiency estimation of histograms is analyzed from ECG signals from database. The entire method has been applied for convenience to different ECG record files taken from MIT-BIH database. Twelve leads are used from multi-lead ECG database which contains a 3600 Hz sampling frequency. The entire algorithm is executed on MATLAB R2014a. In this, the proposed method performance efficiency is evaluated.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
An ECG signal is a most important distinguishing mechanism for recording the electrical movement of heart with the help of ‘P, Q, R, S and T’ segments of signal. So this distinguished waveform provides the fundamental facts about condition of heart patients (Chen et al. 2016). In an ECG signal, ‘QRS’ complex is very important for diagnosis of cardiac abnormalities. These particular units of electrical signal contained the ‘PQ’, ‘ST’, ‘QS’, ‘ST’ and QT segments. The ‘QRS’ complex part shown in ECG signal known as the ‘J’ point (D’Aloia et .al. 2019). The detectable heart sounds are produced by valves of cardiac which are separated or closed with the help of swirling flows. In case of normal or healthy adults, two types of natural heart sounds are audible. It takes place in order of a cardiac phase. The characteristics of ECG signal provides the elementary characteristics like duration and frequency which can be utilized for ECG signal analysis. Several methods are proposed for recognition and detection of QRS complex signal from the ECG signal. The QRS complex signal is obtained for ECG signal detection (Halder et al. 2016).
This paper proposes a novel method which involves minimum statistical detection. By diagnosing the clinically important feature from the ECG signal could be used to recognize the cardiac abnormalities (Lin et al. 2019). The features are recognized through the histogram analysis technique as well as adaptive threshold significance. The ‘R’ peak is distinguished from the first as well as next and accordingly ‘T’ wave and ‘P’ wave have also been identified. In the analysis of ECG signal there is diverse type of noise similar to base line wander noise (Bae and Kwon 2019). The power line noise, movement of object etc., are also integrated. Here power line noise is generated and included in the ECG signal. The maximum detection rate is obtained for P, T and QRS peaks (Wang et al. 2019). The performance analysis of these algorithms is evaluated with fifty, unique concurrently record 12-lead ECG recordings taken from the standard ECG database.
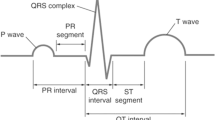
For a normal ECG signal, the noise is included by means of a base line noise. The desired ECG signal follows the sound as well as drift. Subsequent to receiving the ECG dataset, the primary pace is to eliminate the intrinsic sound from the ECG signal (Li et al. 2020). The characteristic of noises, that influence the ECG signal are power line and base line noise of 50/60 Hz. The FIR filtering is utilized to eliminate the 50 Hz power line noise. Subsequent to filtering the signal by the FIR filter, the filtered signals were normal in intermediate filter. At first, of 200 ms filtered ECG signals are stored and extracted. Subsequently its mean has been computed. In the Fig. 1 shows a common ECG signal and its different segments (Shao et al. 2020).
The different intervals of ECG signal is shown in above figure, the ‘RR’ interval involving from ‘R’ wave to the next ‘R’ wave. This is the standard inactive heart time between the 50 and 100 beats per minutes (bpm) and the time interval 0.6–1.2 s. ‘P’ wave is the electrical vector that originates at the ‘SA’ joint to the ‘AV’ node. It is spread starting from the left atrium to the right atrium. This is known as the ‘P’ wave in the ECG signal. The time of ‘P’ wave period is 80 ms. The ‘PR’ time period is calculated since the start of the ‘P’ wave as well as starting point of the ‘QRS’ composite wave. The ‘PR’ period replicates the electrical impulse time that is moved from the ‘AV’ node to ‘SA’ node. The time duration of ‘QRS’ wave 120–200 ms. The ‘PR’ section connect the ‘P’ wave and the ‘QRS’ complex signal. Electrical movements do not produce reduction of straight line and it purely moves downward the ventricles. This shows up direction of the ECG signal. The time duration of ‘PR’ signal time durations are 50–120 ms. The ‘J’ point is the ‘QRS’ complex signal replicate the quick de-polarization of left to the right ventricles (Singh et al. 2018a, b).
The ‘QRS’ complex signal typically have a large amount of amplitude than the P-wave. The time durations are 80–120 ms. The ‘ST’ section connect to the ‘QRS’ composite and the ‘T’ wave signal (Jangra et al. 2020). The ‘ST’ section represents the time period after the ventricles are de-polarized and the time duration 80–120 ms. The ‘T’ wave represents the repolarization of the ventricles. The time duration of ‘T’ signal is 160 ms. The ‘ST’ period is calculated from the ‘J’ position to the ending point of the ‘T’ signal (Lih et al. 2020). The time duration of this ‘T’ wave is 320 ms. The ‘QT’ period is calculated starting point of the ‘QRS’ complex signal to the ending point of the ‘T’ wave. It does vary with the heart rate. The period of ‘QT’ period is 300–430 ms. Table 1 represents the specified frequency range, wave duration for different segments of ECG signal in tabular form.
The desired features of ECG signals are extracted by Mel-frequency cepstral coefficient (MFCC) method, and this method has been reported in this paper. The spectrogram analysis of 10 ECG samples is also presented. The support vector machine (SVM) and ‘k’ nearest neighbor (k-NN) method are also used for detection and recognition of ECG signal (Singh et al. 2018a, b). Usually a threshold technique is used for recognition and identification of ‘R’ peaks in ECG signal. The ECG signal frequency and time interval parameter is extracted from the histogram analysis. It is necessary to improve the QRS detection accuracy. An arithmetical methodology is described for resolving the feature extraction by digital signal approach (Sahu et al. 2020). The remaining part of this manuscript is prepared as follows. In Sect. 2 shows the proposed architecture of ECG signal detection method. In Sects. 3 and 4 feature extraction technique of ECG signal and SVM (Sivaparthipan et al. 2020), k-NN classification methods are discussed. In Sect. 5 discussed the performance result of ECG signal. To conclude in Sect. 6 shows the conclusion part of this study.
2 Related works and proposed system for ECG recognition
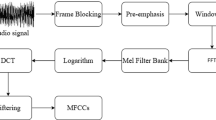
In this section, ECG signal database is taken and the proposed feature extraction technique and their statistical analysis in term of their mean value are presented. After that, the general classification structural design is introduced (Dokur and Olmez 2020). Two machine learning algorithms (SVM and k-NN) have been used for detection. A high pass filter is used to remove the high frequency components present in ECG signal. This proposed method can achieve better detection rate than previous works. The ECG signal detection method has been proposed that use the MFCC’s feature extraction. The detection efficiency outcomes were similar to individuals achieved by ECG signal. The experimental outcome shows that SVM and k-NN give superior detection efficiency rates compared to other approaches shown in Fig. 2. The proposed block diagram is shown in Fig. 2 for ECG detection structure (Martis et al.2012). As representation in block diagram, in the feature extraction, the ECG signal is first processed and is converted it into a feature vectors set. The different feature vectors are used to train the SVM and k-NN classifier. In the other phase, same feature vectors are generated for testing stage for both classifiers. After all training and testing data sets pass through the classifier for the classification output (Yeh et al. 2009).
By this proposed method, the ECG signal is segmented by the peaks. Then, the time period and diastolic phase data had been used for ECG signal detection. This study has three most important goals (Fira and Goras 2008). First from the previous study, the efficiency of the ECG detection rate is observed depending on acoustic features. Second one is based on machine learning algorithms applied to the ECG signal where acoustic features are passed through the SVM and k-NN classifier. In the third case, the histogram analysis of ECG signal is carried out (Lu et al 2000).
Feature extraction using MFCC technique: the feature extraction using MFCC process consist of six operations: (a). pre-emphasis, (b). windowing, (c). Fourier transform, (d). Mel-filter bank, (e). non-linear (log) transformation, (f) discrete cosine transforms (DCT). In the pre-emphasis method the ECG signals are enhanced to minimize the signal distortion. The Hamming window is used to separate a particular signal of frame and maintains the continuity. The Fourier transform procedure is convenient to the windowed signal for conversion from time ___domain to frequency ___domain (spectral) (Singh et al. 2019a, b, c). The feature extraction using MFCC techniques have been more effective in numerous auditory prototype recognition tasks (Laguna et al. 1994) (Fig. 3).
3 The mathematical analysis for MFCC features extraction is given below:
In pre-emphasis phase a first order high pass filter is used. The input signal ‘p[n]’ and pre-emphasis coefficient ‘\(\alpha \)', the values vary between 0.9 to1.0, the input signal in time phase for the filter is articulated as:
The resulting signal is obtained by multiplying the new signal ‘p[n]’, with the window v[n], at time n.
Hamming window is employed for extracting out the MFCC feature coefficients that shift the signal coefficients in the direction of zeros to the margins for avoiding the dissimilarities. Computationally, the Hamming and rectangular window are characterized by:
The simple difficulty involved in FFT is just finding the individual coefficients of ‘M’. That are the power of ‘2’ precisely, DFT can be analyzed as the frequency ___domain analysis shown in Fig. 4:
They overlap next to the ___location of the margin of every filter. A usual method for attaining these values is as following:
The exceeding shape contains twenty triangular band-pass filters. The filter is located at normal duration beside the Mel-scale frequency, which is articulated as (Singh et al. 2019c):
At this point, DCT is used to convert it from time ___domain to cepstrum ___domain. Computationally, the MFCCs could be represented as:
The acoustic features using MFCC varies with the duration of time, arithmetical moment is removed from the auditory vectors by the related interval. At this point, ‘p(n)’ is a input speech signal with ‘N’ frame, those MFCC vectors are indicated by ‘vij’ where ‘j’ represent the characteristic element and ‘i’ depict the number of frames, and it is articulated as:
After this, two different types of arithmetical moments are used. Initially the mean 'Ej’ of every MFCC feature ‘Uj’ is extracted. The mean ‘E’ calculated from each sample is given as:
4 SVM and k-NN classifier for proposed system
Training and testing data for SVM classifier taken as example (Sivaparthipan et al. 2020):
The dataset taken is (x1, y1)…(xn, yn)
x1 is a set of training feature vector, y1 is corresponding testing feature vectors.
Each experiment i:
xi is feature is present or not present, here shows xi(j) is real value
‘w’ is the weight of each vector, which is the best linear separator. And ‘x’ is the feature vector
Deciding the margin of each vector:
Let line L: w*x + c = 0
w(1)x(1) + w(2)x(2) + b = 0
At point ‘A’ is data set point A = (XA(1), XA(2))
Let ‘M’ is the arbitrary point in hyper plane:
Point ‘M’ on a line = (Xm(1), Xm(2))
SVM defines the hyper plane with closest distance
w* = arg max [min dh (α(Xn)]
for proper classification:
yn[wT α(x) + b] = {≥ 0 Correct, < 0 incorrect
w* = arg max[min dH (α(xn))]
Distance of closest point to ‘x’.
min ½ |w2|
yn [wT α (xn) + b] ≥ 1 primal form of SVM
k- NN classifier (Yeh et al. 2009):
(x1, y1)…(xn, yn)
xi € Rd, yi € {0, 1}
‘xi’ known as arbitrary data and ‘yi’ known as binary data
The distance matrix:
xi = (xi1, xi2…xid)
by using probabilistic approach
Random variable ‘y ~ p’, p(y) = fraction of point
Nk(x) nearest point to x
5 Performance evaluation and experimental results
In this experimental work, the extraction process of features is provided in Sect. 3. The classification mechanism using SVM and k-NN classification model is derived in Sect. 4. To categorize and estimate the detection efficiency of the proposed SVM and k-NN classifiers are compared with the log regression (LR), deep neural network (DNN) and Gaussian mixture model (GMM) classifiers for recognition of ECG signal. To appraise the presentation of the proposed technique, 12-dissimilar leads of ECG signal are considered from the ECG database. The algorithms are accomplished to recognize the ‘R’ peak completely. Three arithmetical calculations are used to find the presence of ‘R’ peak and to calculate its metrics. These are detection accuracy (DA), true positives (TP) the relative amount of the number of appropriately recognized peaks and sensitivity (Se), the metrics are specified by:
where false negatives (FN) is the quantity of missed trials. The positive prognostic accurateness (+ P), is the relation of the amount of correctly recognized trials (TP), to the sum of measures of detected peaks by the analyzer and it is measured by:
where false positives (FP) is the amount of incorrectly recognized trials. An additional performance computation is DA calculated by the ratio of detected peaks and the total number of peaks.
Table 2 show the detection accuracy (DA %), positive peak predictivity (+ P %) and sensitivity (Se %), of different ‘R’ peaks of various ECG data files. Consequently proposed method is compared with other methods. Furthermore, proposed method comparatively does not include several statistical computations.
The proposed technique is applied to the ECG database accessed from MIT-BIH Arrhythmia and performance metrics are evaluated. That consists by addition of standardized amount of sound to fresh ECG recording with the help of MIT-BIH database. With the intention of estimate the critical cardiac condition, simply the records from the database (file no. 100–118) are utilized for this observation.
Figures 5, 6 and 7 represent the ECG signal waveform and their respective spectrograms. A spectrogram analysis of a signal is frequently used to show, how much frequencies are present in a sequential ECG signal that fluctuate with time. To represent the spectrogram presented in Figs. 5, 6 and 7 the sampling frequency obtained is 3.6 kHz.
ECG signal plot and spectrogram analysis a ECG signal of 100 m database b spectrogram analysis of 100 m ECG signal c ECG signal of 103 m database d spectrogram analysis of 103 m ECG signal e ECG signal of 105 m database f spectrogram analysis of 105 m ECG signal g ECG signal of 108 m database h spectrogram analysis of 108 m ECG signal
The frame size has been placed to 20 ms of signal, through a 10 ms overlie. In this spectrogram, it is observed that the distinguished features of the ECG signal are mostly concentrated in the low frequency region. With the clean ECG signal waveform, every essential characteristic of ECG is extracted. The recognition of the ‘QRS’ complex in ECG signal is the primary task and a large amount of this significant factor is used for feature extraction. The recognized ‘R’ peak is estimated for every strike of the waveform.
From the position of ‘R’ peak, the other fudicial points on the signal are detected. Consequently to identify a precise ‘QRS’ complex signal is a significant job in ECG study. Subsequent to effective recognition of ‘R’ crest, the discernible histogram contained could be removed in the histogram. The histograms of different ECG signal are shown in Figs. 8 and 9. After executing the whole procedure for each ECG signal, acoustic feature can be detected from each ECG signal by computing mean using MFCCs. Figure 10 shows the mean values of MFCC of 10 ECG signals respectively. Every individual ECG signal have exceptional acoustic features.
The acoustic coefficients are calculated in terms of mean, 100 m files to 118 m files ECG signal using MFCC feature extraction. The different ECG file signal mean values are shown in Fig. 10.
The normal recognition of detection efficiency of ECG signal consists of a testing stage and training stage. For training the data, initially an ECG database is taken where all ECG signals are presented. Depending on testing and training value, classifiers calculate the preferred value of the train data by which the test data features are recognized. Subsequently the features vector is computed for every ECG signal model in the training module. Following these steps, the next procedure is to extract the features in testing section. In the extraction of particular ECG signal features of different classes are compared in Table 3.
Table 3 shows the result of different classifiers detection efficiency rates. SVM and k-NN have extensively superior detection rates than all existing classifiers. Proposed results on SVM and k-NN might be contradictory with some other estimations connecting to GMM, LR and DNN. At this time pre-eminence of SVM and k-NN are shown above the other learning algorithms as it reached an outstanding detection efficiency of 95.45% and 96.57% respectively.
6 Conclusion
In this article, acoustic features of ECG signals are extracted using MFCC feature extraction for recognizing the ECG signal and using SVM and k-NN classifiers, the detection efficiency is evaluated. This article have center of attention of finding the detection efficiency performance based on the acoustic features of ECG signal. Identification and Detection of dissimilar model of ECG signal by their graphical illustration verification of 12 lead ECG waveforms are explained in this article. The uniqueness of this methodology is computation of histogram by 20 ms different section value on ECG signal for the recognition of ‘R’ peaks. The detection efficiency obtained is 95.45% and 96.57% from SVM and k-NN classifiers respectively using the proposed technique for ECG signal feature extraction and classification. This technique is appropriate to be used in the analysis of similar structural design for real time applications. The technical flaw using this method is connected to the resolution and sampling rate of the ECG signal.
Change history
28 November 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s12652-024-04937-1
References
Bae TW, Kwon KK (2019) Efficient real-time R and QRS detection method using a pair of derivative filters and max filter for portable ECG device. ApplSci 9(19):4128. https://doi.org/10.3390/app9194128
Chen TE, Yang SI, Ho LT, Tsai KH, Chen YH, Chang YF, Lai YH, Wang SS, Tsao Y, Wu CC (2016) S1 and S2 heart sound recognition using deep neural networks. IEEE Trans Biomed Eng 64(2):372–380. https://doi.org/10.1109/tbme.2016.2559800
D’Aloia M, Longo A, Rizzi M (2019) Noisy ECG signal analysis for automatic peak detection. Information 10(2):35. https://doi.org/10.3390/info.10020035
Dokur Z, Ölmez T (2020) Heartbeat classification by using a convolutional neural network trained with Walsh functions. Neural ComputAppl. https://doi.org/10.1007/s00521-020-04709-w
Fira CM, Goras L (2008) An ECG signals compression method and its validation using NNs. IEEE Trans Biomed Eng 55(4):1319–1326. https://doi.org/10.1109/tbme.2008.918465
Halder B, Mitra S, Mitra M (2016) Detection and identification of ECG waves by histogram approach. In: 2016 2nd international conference on control, instrumentation, energy & communication (CIEC) 2016, IEEE, pp 168–172. https://doi.org/10.1109/ciec.2016.7513749
Jangra M, Dhull SK, Singh KK (2020) ECG arrhythmia classification using modified visual geometry group network (mVGGNet). J Intell Fuzzy Syst. https://doi.org/10.3233/jifs-191135
Laguna P, Jané R, Caminal P (1994) Automatic detection of wave boundaries in multilead ECG signals: validation with the CSE database. Comput Biomed Res 27(1):45–60. https://doi.org/10.1006/cbmr.1994.1006
Li H, Wei X, Zuo S, Dou Q, Ding M, Cao L, Gong Z, Wang R, Chen X, Wang B, Prades JD (2020) Arrhythmia classification algorithm based on multi-feature and multi-type optimized SVM. Am Sci Res J EngTechnolSci 63(1):72–86
Lih OS, Jahmunah V, San TR, Ciaccio EJ, Yamakawa T, Tanabe M, Kobayashi M, Faust O, Acharya UR (2020) Comprehensive electrocardiographic diagnosis based on deep learning. Artif Intell Med 103: https://doi.org/10.1016/j.artmed.2019.101789
Lin WH, Ji N, Wang L, Li G (2019) A characteristic filtering method for pulse wave signal quality assessment. In: 2019 41st annual international conference of the IEEE engineering in medicine and biology society (EMBC) 2019. IEEE, Berlin, pp 603–606. https://doi.org/10.1109/EMBC.2019.8856811.
Lu Z, Kim DY, Pearlman WA (2000) Wavelet compression of ECG signals by the set partitioning in hierarchical trees algorithm. IEEE Trans Biomed Eng 47(7):849–856. https://doi.org/10.1109/10.846678
Martis RJ, Acharya UR, Mandana KM, Ray AK, Chakraborty C (2012) Application of principal component analysis to ECG signals for automated diagnosis of cardiac health. Expert SystAppl 39(14):11792–11800. https://doi.org/10.1016/J.cswa.2012.04.072
Sahu N, Peng D, Sharif H (2020) An innovative approach to integrate unequal protection-based steganography and progressive transmission of physiological data. SN ApplSci 2(2):237. https://doi.org/10.1007/s42452-020-1992-0
Shao M, Zhou Z, Bin G, Bai Y, Wu S (2020) A wearable electrocardiogram telemonitoring system for atrial fibrillation detection. Sensors 20(3):606. https://doi.org/10.3390/s20030606
Singh MK, Singh AK, Singh N (2018a) Acoustic comparison of electronics disguised voice using different semitones. Int J EngTechnol (UAE) 7(2):98. https://doi.org/10.14419/ijet.v7i2.16.11502
Singh MK, Singh AK, Singh N (2018b) Disguised voice with fast and slow speech and its acoustic analysis. Int J Pure Appl Math 118(14):241–246
Singh MK, Singh AK, Singh N (2019a) Multimedia analysis for disguised voice and classification efficiency. Multimed Tools Appl 78(20):29395–29411. https://doi.org/10.1007/s11042-018-6718-6
Singh M, Nandan D, Kumar S (2019b) Statistical analysis of lower and raised pitch voice signal and its efficiency calculation. Traitement du Signal 36(5):455–461. https://doi.org/10.18280/ts.360511
Singh MK, Singh N, Singh AK (2019c) Speaker's voice characteristics and similarity measurement using Euclidean distances. In: 2019 international conference on signal processing and communication (ICSC). IEEE, pp 317–322. https://doi.org/10.1109/icsc.45622.2019.8938366
Sivaparthipan CB, Karthikeyan N, Karthik S (2020) Designing statistical assessment healthcare information system for diabetics analysis using big data. Multimed Tools Appl 79:8431–8444. https://doi.org/10.1007/s11042-018-6648-3
Wang J, Wang P, Wang S (2019) Automated detection of atrial fibrillation in ECG signals based on wavelet packet transform and correlation function of random process. Biomed Signal Process Control 55:101662. https://doi.org/10.1016/j.bspc.2019.101662
Yeh YC, Wang WJ, Chiou CW (2009) Cardiac arrhythmia diagnosis method using linear disciminant analysis on ECG signals. Measurement 42(5):778–89. https://doi.org/10.1016/j.measurement.2009.01.004
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s12652-024-04937-1
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Arpitha, Y., Madhumathi, G.L. & Balaji, N. RETRACTED ARTICLE: Spectrogram analysis of ECG signal and classification efficiency using MFCC feature extraction technique. J Ambient Intell Human Comput 13, 757–767 (2022). https://doi.org/10.1007/s12652-021-02926-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-021-02926-2