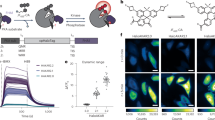
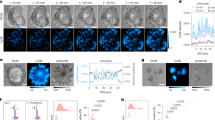
Abstract
The ability to explore cell signalling and cell-to-cell communication is essential for understanding cell biology and developing effective therapeutics. However, it is not yet possible to monitor the interaction of cells with their environments in real time. Here, we show that a fluorescent sensor attached to a cell membrane can detect signalling molecules in the cellular environment. The sensor is an aptamer (a short length of single-stranded DNA) that binds to platelet-derived growth factor (PDGF) and contains a pair of fluorescent dyes. When bound to PDGF, the aptamer changes conformation and the dyes come closer to each other, producing a signal. The sensor, which is covalently attached to the membranes of mesenchymal stem cells, can quantitatively detect with high spatial and temporal resolution PDGF that is added in cell culture medium or secreted by neighbouring cells. The engineered stem cells retain their ability to find their way to the bone marrow and can be monitored in vivo at the single-cell level using intravital microscopy.
This is a preview of subscription content, access via your institution
Access options
Access to this article via ICE Institution of Civil Engineers is not available.
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Halin, C., Mora, J., Sumen, C. & von Andrian, U. In vivo imaging of lymphocyte trafficking. Annu. Rev. Cell. Dev. Biol. 21, 581–603 (2005).
Karp, J. M. & Teo, G. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell 4, 206–216 (2009).
Ferreira, L., Karp, J. M., Nobre, L. & Langer, R. New opportunities: the use of nanotechnologies to manipulate and track stem cells. Cell Stem Cell 3, 136–146 (2008).
Laughlin, S. T., Baskin, J. M., Amacher, S. L. & Bertozzi, C. R. In vivo imaging of membrane-associated glycans in developing zebrafish. Science 320, 664–667 (2008).
Cook, B. N. & Bertozzi, C. R. Chemical approaches to the investigation of cellular systems. Bioorg. Med. Chem. 10, 829–840 (2002).
Giepmans, B. N. G., Adams, S. R., Ellisman, M. H. & Tsien, R. Y. The fluorescent toolbox for assessing protein ___location and function. Science 312, 217–224 (2006).
Hoffmann, C. et al. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nature Methods 2, 171–176 (2005).
Pollok, B. A. & Heim, R. Using GFP in FRET-based applications. Trends Cell Biol. 9, 57–60 (1999).
Modi, S. et al. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nature Nanotech. 4, 325–330 (2009).
Albizu, L. et al. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nature Chem. Biol. 6, 587–594 (2010).
Rider, T. et al. A B cell-based sensor for rapid identification of pathogens. Science 301, 213–215 (2003).
Beigi, R., Kobatake, E., Aizawa, M. & Dubyak, G. Detection of local ATP release from activated platelets using cell surface-attached firefly luciferase. Am. J. Physiol. Cell Physiol. 276, C267–C278 (1999).
Orynbayeva, Z. et al. Visualization of membrane processes in living cells by surface-attached chromatic polymer patches. Angew. Chem. Int. Ed. 44, 1092–1096 (2005).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Ellington, A. & Szostak, J. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Liu, J., Cao, Z. & Lu, Y. Functional nucleic acid sensors. Chem. Rev. 109, 1948–1998 (2009).
Zhao, W., Brook, M. & Li, Y. Design of gold nanoparticle based colorimetric biosensing assays. ChemBioChem 9, 2363–2371 (2008).
Sefah, K. et al. Nucleic acid aptamers for biosensors and bio-analytical applications. Analyst 134, 1765–1775 (2009).
Nutiu, R. & Li, Y. In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. 44, 1061–1065 (2005).
Keefe, A. D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nature Rev. Drug Discov. 9, 573–550 (2010).
Fang, X. & Tan, W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc. Chem. Res. 43, 48–57.
Dhar, S., Kolishetti, N., Lippard, S. J. & Farokhzad, O. C. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc. Natl Acad. Sci. USA 108, 1850–1855.
Ankrum, J. & Karp, J. M. Mesenchymal stem cell therapy: two steps forward, one step back. Trends Mol. Med. 16, 203–209 (2010).
López Ponte, A. et al. The in vitro migration capacity of human bone marrow mesenchymal stem cells: comparison of chemokine and growth factor chemotactic activities. Stem Cells 25, 1737–1745 (2007).
Beckermann, B. et al. VEGF expression by mesenchymal stem cells contributes to angiogenesis in pancreatic carcinoma. Br. J. Cancer 99, 622–631 (2008).
Ball, S. G., Shuttleworth, C. A. & Kielty, C. M. Mesenchymal stem cells and neovascularization: role of platelet-derived growth factor receptors. J. Cell. Mol. Med. 11, 1012–1030 (2007).
Fang, X., Sen, A., Vicens, M. & Tan, W. Synthetic DNA aptamers to detect protein molecular variants in a high-throughput fluorescence quenching assay. ChemBioChem 4, 829–834 (2003).
Vicens, M., Sen, A., Vanderlaan, A., Drake, T. & Tan, W. Investigation of molecular beacon aptamer-based bioassay for platelet-derived growth factor detection. ChemBioChem 6, 900–907 (2005).
Green, L. S. et al. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry 35, 14413–14424 (1996).
Adams, G. B. et al. Haematopoietic stem cells depend on Gα(s)-mediated signalling to engraft bone marrow. Nature 459, 103–107 (2009).
Lo Celso, C. et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457, 92–97 (2009).
Lo Celso, C., Wu, J. W. & Lin, C. P. In vivo imaging of hematopoietic stem cells and their microenvironment. J. Biophoton. 2, 619–631 (2009).
Sarkar, D. et al. Chemical engineering of mesenchymal stem cells to induce a cell rolling response. Bioconjug. Chem. 19, 2105–2109 (2008).
Sarkar, D. et al. Engineered mesenchymal stem cells with self-assembled vesicles for systemic cell targeting. Biomaterials 31, 5266–5274 (2010).
Zhao, W. et al. Mimicking the inflammatory cell adhesion cascade by nucleic acid aptamer programmed cell–cell interactions. FASEB J. doi:10.1096/fj.10-178384 (2011).
Lai, R. Y., Plaxco, K. W. & Heeger, A. J. Aptamer-based electrochemical detection of picomolar platelet-derived growth factor directly in blood serum. Anal. Chem. 79, 229–233 (2007).
Leitzel, K. et al. Elevated plasma platelet-derived growth factor B-chain levels in cancer patients. Cancer. Res. 51, 4149–4154 (1991).
Ogunniyi, A. O., Story, C. M., Papa, E., Guillen, E. & Love, J. C. Screening individual hybridomas by microengraving to discover monoclonal antibodies. Nature Protoc. 4, 767–782 (2009).
Au, P. et al. Paradoxical effects of PDGF-BB overexpression in endothelial cells on engineered blood vessels in vivo. Am. J. Pathol. 175, 294–302 (2009).
Mazo, I. B. et al. Hematopoietic progenitor cell rolling in bone marrow microvessels: parallel contributions by endothelial selectins and vascular cell adhesion molecule 1. J. Exp. Med. 188, 465–474 (1998).
Jayasena, S. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45, 1628–1650 (1999).
Cox, J. C., Rudolph, P. & Ellington, A. D. Automated RNA selection. Biotechnol. Prog. 14, 845–850 (1998).
Lou, X. et al. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl Acad. Sci. USA 106, 2989–2994 (2009).
Kim, G., Kim, K., Oh, M. & Sung, Y. The detection of platelet derived growth factor using decoupling of quencher-oligonucleotide from aptamer/quantum dot bioconjugates. Nanotechnology 20, 175503 (2009).
Tang, Z. et al. Aptamer switch probe based on intramolecular displacement. J. Am. Chem. Soc. 130, 11268–11269 (2008).
Hui, E. & Bhatia, S. Micromechanical control of cell–cell interactions. Proc. Natl Acad. Sci. USA 104, 5722–5726 (2007).
Zhao, W., Gao, Y., Kandadai, S. A., Brook, M. A. & Li, Y. DNA polymerization on gold nanoparticles through rolling circle amplification: towards novel scaffolds for three-dimensional periodic nanoassemblies. Angew. Chem. Int. Ed. 45, 2409–2413 (2006).
Hayashi, S., Hazama, A., Dutta, A. K., Sabirov, R. Z. & Okada, Y. Detecting ATP release by a biosensor method. Sci. STKE 2004, pl14 (2004).
Zhao, W., Teo, G., Kumar, N. & Karp, J. Chemistry and material science at the cell surface. Mater. Today 13, 14–21 (2010).
Acknowledgements
The authors thank Lei Xu and Dannie Wang for the preparation of PDGF-producing MDA-MB-231 cells. This work was supported by the National Institutes of Health (NIH; grants nos. HL097172, HL095722 and DE019191 to J.M.K., grants nos. HL082792 and NS059348 to J.L., and grant no. NIAID 5RC1AI086152 to J.C.L.), by the Charles A. Dana Foundation (J.C.L.) and by the American Heart Association (grant no. 0970178N to J.M.K.). W.Z. is supported by an International Human Frontier Science Program Organization postdoctoral fellowship. Y.J.Y. holds a National Science Foundation Graduate Fellowship.
Author information
Authors and Affiliations
Contributions
W.Z. and J.M.K. are responsible for study concept and design. W.Z., J.M.K., J.L., J.C.L., C.P.L., D.S. and R.K. prepared the manuscript. W.Z., S.S., J.C., Y.J.Y., M.L.L., S.B., A.L.C., J.A.P, W.T., I.A.D. and C.C. carried out experiments and performed data analysis. R.K.J. provided genetically engineered cells.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 874 kb)
Rights and permissions
About this article
Cite this article
Zhao, W., Schafer, S., Choi, J. et al. Cell-surface sensors for real-time probing of cellular environments. Nature Nanotech 6, 524–531 (2011). https://doi.org/10.1038/nnano.2011.101
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2011.101