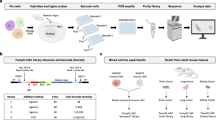
Abstract
A major challenge in neuronal stem cell biology lies in characterization of lineage-specific reprogrammed human neuronal cells, a process that necessitates the use of an assay sensitive to the single-cell level. Single-cell gene profiling can provide definitive evidence regarding the conversion of one cell type into another at a high level of resolution. The protocol we describe uses Fluidigm Biomark dynamic arrays for high-throughput expression profiling from single neuronal cells, assaying up to 96 independent samples with up to 96 quantitative PCR (qPCR) probes (equivalent to 9,216 reactions) in a single experiment, which can be completed within 2–3 d. The protocol enables simple and cost-effective profiling of several hundred transcripts from a single cell, and it could have numerous utilities.
This is a preview of subscription content, access via your institution
Access options
Access to this article via ICE Institution of Civil Engineers is not available.
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dolmetsch, R. & Geschwind, D.H. The human brain in a dish: the promise of iPSC-derived neurons. Cell 145, 831–834 (2011).
Li, H.H. et al. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 335, 414–417 (1988).
Cauli, B. et al. Molecular and physiological diversity of cortical nonpyramidal cells. J. Neurosci. 17, 3894–3906 (1997).
Cauli, B. & Lambolez, B. in Unravelling Single Cell Genomics 81–92 (The Royal Society of Chemistry, 2010).
Koirala, S. & Corfas, G. Identification of novel glial genes by single-cell transcriptional profiling of Bergmann glial cells from mouse cerebellum. PLoS One 5, e9198 (2011).
Lambolez, B. et al. AMPA receptor subunits expressed by single Purkinje cells. Neuron 9, 247–258 (1992).
Surmeier, D.J. et al. Dopamine receptor subtypes colocalize in rat striatonigral neurons. Proc. Natl. Acad. Sci. USA 89, 10178–10182 (1992).
Tietjen, I., Rihel, J. & Dulac, C.G. Single-cell transcriptional profiles and spatial patterning of the mammalian olfactory epithelium. Int. J. Dev. Biol. 49, 201–207 (2005).
Tietjen, I. et al. Single-cell transcriptional analysis of neuronal progenitors. Neuron 38, 161–175 (2003).
Mackler, S.A., Brooks, B.P. & Eberwine, J.H. Stimulus-induced coordinate changes in mRNA abundance in single postsynaptic hippocampal CA1 neurons. Neuron 9, 539–548 (1992).
Mackler, S.A. & Eberwine, J.H. Diversity of glutamate receptor subunit mRNA expression within live hippocampal CA1 neurons. Mol. Pharmacol. 44, 308–315 (1993).
Ginsberg, S.D. et al. Single-cell gene expression analysis: implications for neurodegenerative and neuropsychiatric disorders. Neurochem. Res. 29, 1053–1064 (2004).
Sucher, N.J. & Deitcher, D.L. PCR and patch-clamp analysis of single neurons. Neuron 14, 1095–1100 (1995).
Lao, K.Q. et al. mRNA-sequencing whole transcriptome analysis of a single cell on the SOLiD system. J. Biomol. Tech. 20, 266–271 (2009).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
White, A.K. et al. High-throughput microfluidic single-cell RT-qPCR. Proc. Natl. Acad. Sci. 108, 13999–14004 (2011).
Spurgeon, S.L., Jones, R.C. & Ramakrishnan, R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One 3, e1662 (2008).
Pang, Z.P. et al. Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 (2011).
Yoo, A.S. et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 (2011).
Narsinh, K.H. et al. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J. Clin. Invest. 121, 1217–1221 (2011).
Liss, B. & Roeper, J. Correlating function and gene expression of individual basal ganglia neurons. Trends Neurosci. 27, 475–481 (2004).
Weng, J.Y., Lin, Y.C. & Lien, C.C. Cell type-specific expression of acid-sensing ion channels in hippocampal interneurons. J. Neurosci. 30, 6548–6558 (2010).
McClung, C.A. & Nestler, E.J. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology 33, 3–17 (2008).
Lammel, S., Ion, D.I., Roeper, J. & Malenka, R.C. Projection-specific modulation of dopamine neuron synapses by aversive and rewarding stimuli. Neuron 70, 855–862 (2011).
Lobo, M.K. et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science 330, 385–390 (2010).
Allen, S.E., Darnell, R.B. & Lipscombe, D. The neuronal splicing factor Nova controls alternative splicing in N-type and P-type CaV2 calcium channels. Channels (Austin) 4, 483–489 (2010).
Bharadwaj, R. & Kolodkin, A.L. Descrambling Dscam diversity. Cell 125, 421–424 (2006).
Hodne, K., Haug, T.M. & Weltzien, F.A. Single-cell qPCR on dispersed primary pituitary cells—an optimized protocol. BMC Mol. Biol. 11, 82 (2010).
Morris, J., Singh, J.M. & Eberwine, J.H. Transcriptome analysis of single cells. J. Vis. Exp. published online, doi:10.3791/2634 (2011).
Esumi, S., Kaneko, R., Kawamura, Y. & Yagi, T. Split single-cell RT-PCR analysis of Purkinje cells. Nat. Protoc. 1, 2143–2151 (2006).
Li, Y. et al. An improved one-tube RT-PCR protocol for analyzing single-cell gene expression in individual mammalian cells. Anal. Bioanal. Chem. 397, 1853–1859 (2010).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Bustin, S.A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000).
Thornton, B. & Basu, C. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 39, 145–154 (2011).
Acknowledgements
A.C. acknowledges the generous support of the AXA Research Fund. Z.P.P. is supported by the Brain and Behavior Research Foundation (National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Award) and the Robert Wood Johnson Foundation. We thank N. Yang for help in experimental work. We thank S. Chavez for initial instruction in the use of the Fluidigm Biomark system, and R. Reijo Pera for access to the Fluidigm Biomark system in her lab. We also thank members of the Malenka and Südhof labs for their comments on the manuscript.
Author information
Authors and Affiliations
Contributions
A.C. and Z.P.P. conceived the protocol, collected the data, presented the figures and wrote the manuscript. T.C.S., M.W. and R.C.M. supervised the project and contributed to the writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Citri, A., Pang, Z., Südhof, T. et al. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat Protoc 7, 118–127 (2012). https://doi.org/10.1038/nprot.2011.430
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.430