Abstract
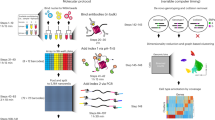
Hi-C is a powerful method that provides pairwise information on genomic regions in spatial proximity in the nucleus. Hi-C requires millions of cells as input and, as genome organization varies from cell to cell, a limitation of Hi-C is that it only provides a population average of genome conformations. We developed single-cell Hi-C to create snapshots of thousands of chromatin interactions that occur simultaneously in a single cell. To adapt Hi-C to single-cell analysis, we modified the protocol to include in-nucleus ligation. This enables the isolation of single nuclei carrying Hi-C–ligated DNA into separate tubes, followed by reversal of cross-links, capture of biotinylated ligation junctions on streptavidin-coated magnetic beads and PCR amplification of single-cell Hi-C libraries. The entire laboratory protocol can be carried out in 1 week, and although we have demonstrated its use in mouse T helper (TH1) cells, it should be applicable to any cell type or species for which standard Hi-C has been successful. We also developed an analysis pipeline to filter noise and assess the quality of data sets in a few hours. Although the interactome maps produced by single-cell Hi-C are sparse, the data provide useful information to understand cellular variability in nuclear genome organization and chromosome structure. Standard wet and dry laboratory skills in molecular biology and computational analysis are required.
This is a preview of subscription content, access via your institution
Access options
Access to this article via Institution of Civil Engineers Library is not available.
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Fraser, P. & Bickmore, W. Nuclear organization of the genome and the potential for gene regulation. Nature 447, 413–417 (2007).
Gibcus, J.H. & Dekker, J. The hierarchy of the 3D genome. Mol. Cell 49, 773–782 (2013).
De Wit, E. & de Laat, W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 (2012).
Ethier, S.D., Miura, H. & Dostie, J. Discovering genome regulation with 3C and 3C-related technologies. Biochim. Biophys. Acta 1819, 401–410 (2012).
Tanay, A. & Cavalli, G. Chromosomal domains: epigenetic contexts and functional implications of genomic compartmentalization. Curr. Opin. Genet. Dev. 23, 197–203 (2013).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Dixon, J.R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Nora, E.P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Dekker, J., Marti-Renom, M.A. & Mirny, L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Furlan-Magaril, M., Várnai, C., Nagano, T. & Fraser, P. 3D genome architecture from populations to single cells. Curr. Opin. Genet. Dev. 31, 36–41 (2015).
Rapkin, L.M., Anchel, D.R.P., Li, R. & Bazett-Jones, D.P. A view of the chromatin landscape. Micron 43, 150–158 (2012).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Nagano, T. et al. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 16, 175 (2015).
Rao, S.S.P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Sexton, T. et al. Sensitive detection of chromatin coassociations using enhanced chromosome conformation capture on chip. Nat. Protoc. 7, 1335–1350 (2012).
Dryden, N.H. et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by capture Hi-C. Genome Res. 24, 1854–1868 (2014).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Tolhuis, B., Palstra, R.J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465 (2002).
Simonis, M., Kooren, J. & de Laat, W. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods 4, 895–901 (2007).
Kilkenny, C., Browne, W.J., Cuthill, I.C., Emerson, M. & Altman, D.G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8, e1000412 (2010).
Krueger, F., Andrews, S.R. & Osborne, C.S. Large scale loss of data in low-diversity illumina sequencing libraries can be recovered by deferred cluster calling. PLoS ONE 6, e16607 (2011).
Acknowledgements
The authors thank I. Clay, N. Cope and K. Tabbada for help and support. This work was supported by the Biotechnology and Biological Sciences Research Council, UK.
Author information
Authors and Affiliations
Contributions
T.N. and P.F. developed and optimized the protocol. Y.L., E.Y. and A.T. contributed to protocol optimization by developing the processing pipeline for single-cell Hi-C and analyzing the sequence data. S.W.W. contributed to optimize the pipeline. W.D. contributed the single-cell isolation procedure. T.N., S.W.W. and P.F. wrote the manuscript, with inputs from all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Nagano, T., Lubling, Y., Yaffe, E. et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc 10, 1986–2003 (2015). https://doi.org/10.1038/nprot.2015.127
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.127