Stanley Institute for Cognitive Genomics
The Stanley Institute for Cognitive Genomics is dedicated to genetics and neuroscience research that will enhance our understanding of schizophrenia, bipolar disorder, major depression and other cognitive disorders. The goal is to improve the diagnosis and treatment of these disorders.
Work at the Stanley Institute for Cognitive Genomics at CSHL is funded by gifts from Theodore and Vada Stanley and grants from the
National Institute of Mental Health.
 Schizophrenia, bipolar disorder and major recurrent depression are cognitive disorders that create an enormous burden on patients, their families and our health care system.
Schizophrenia, bipolar disorder and major recurrent depression are cognitive disorders that create an enormous burden on patients, their families and our health care system.
Because vulnerability to these disorders tends to run in families, genetics is a factor; however the specific genetic components involved and how they impact symptoms or treatments of these disorders has yet to be fully worked out. With recent advances in genomic technologies, CSHL is now poised to unravel the genetic complexity of cognitive disorders. Simultaneously, advanced technologies in neuroscience are allowing CSHL researchers to understand how the brain assembles neural circuits to control behaviors and cognitive processes like attention and decision-making. At the CSHL Stanley Institute for Cognitive Genomics, these two approaches – genetics and neuroscience – are integrated to form a dual-strategy aimed at improving the diagnosis and treatment of schizophrenia, bipolar disorder, depression and other cognitive disorders.
Detailed images of NMDA receptor show how zinc and a potential drug affect its function
December 1, 2016
Drugs precisely targeting portions of the NMDA receptor may have applications in Alzheimer’s, depression and schizophrenia.
Was it better or worse than you expected? Your basal ganglia know – so you can act accordingly
September 21, 2016
Researchers have uncovered a neural circuit that processes evaluations and have succeeded in identifying its sources.
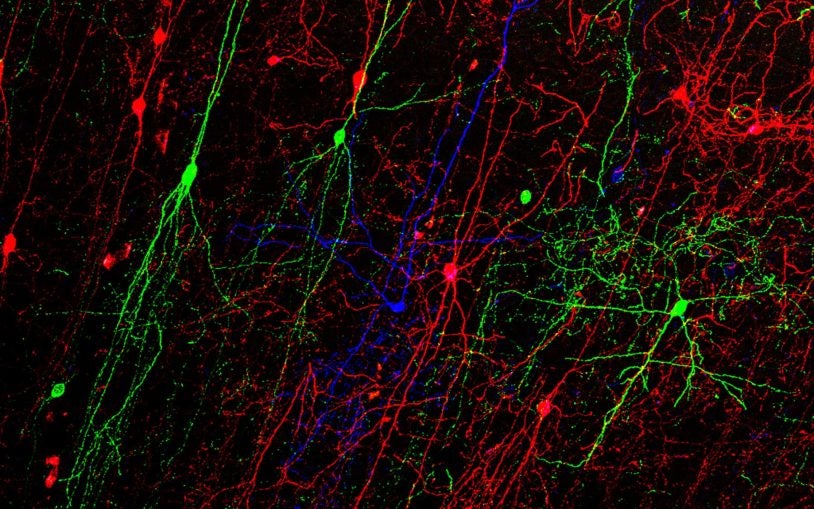
Innovative tools will shed clarifying light on inhibitory neurons
September 21, 2016
Researchers at Cold Spring Harbor Laboratory created a set of tools that combine genetic markers and virus particles to target GABA cell types.
Our brain uses statistics to calculate confidence, make decisions
May 4, 2016
The brain produces feelings of confidence that inform decisions the same way statistics pulls patterns out of noisy data.
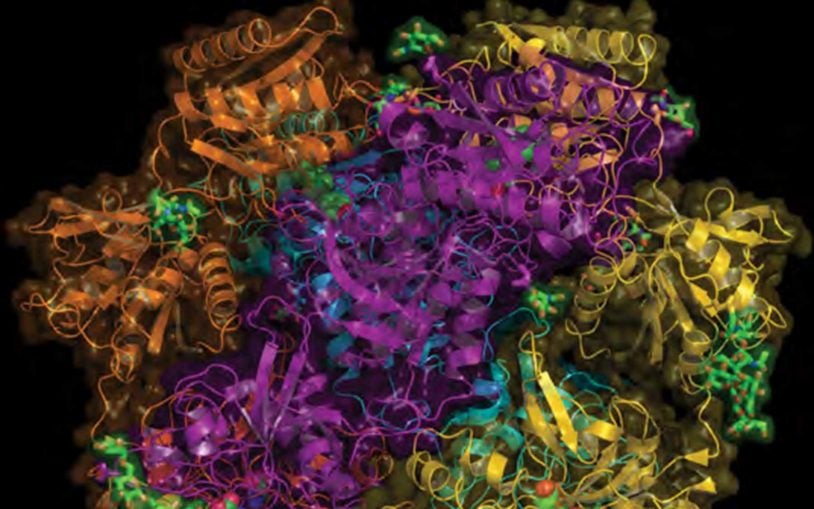
First structural views of the NMDA receptor in action will aid drug development
May 2, 2016
Structural biologists obtained snapshots of the activation of an important type of brain-cell receptor.
CSHL scientists Bo Li and Je Lee win HFSP Research Grant awards
April 12, 2016
Associate Professor Bo Li and Assistant Professor Je Lee, are among 25 teams that have won Human Frontier Science Program Research Program Grants
Discovery of a new X-linked intellectual disability syndrome is aided by web communication tools
December 3, 2015
A geneticist uses social media and online tools to help confirm, document and describe in precise clinical detail a new genetic syndrome in young
Surprised? Cholinergic neurons send brain-wide broadcasts enabling us to learn from the unexpected
August 25, 2015
Researchers find dedicated neurons that rapidly informing multiple subregions in the brain of any surprising rewards or punishments.
Drs. Kepecs and Li honored with 2015 NARSAD Independent Investigator grant awards
May 12, 2015
Dr. Kepecs and Dr. Li received two-year grants to fund their important research.
Scientists discover important communication mechanism between two brain areas implicated in schizophrenia
April 7, 2015
Researchers discover an inhibitory connection between certain brain areas that can control the timing of information flow to the prefrontal cortex
A new brain circuit that controls fear is identified
January 19, 2015
Researchers discover a pathway in that mouse brain that regulates fear memory and behavior
Imaging method linking brainwide cell activation & behavior shows what it means for mice to have sex in mind
January 5, 2015
Automated method detects activity of neurons during specific behaviors, brainwide, at cellular resolution
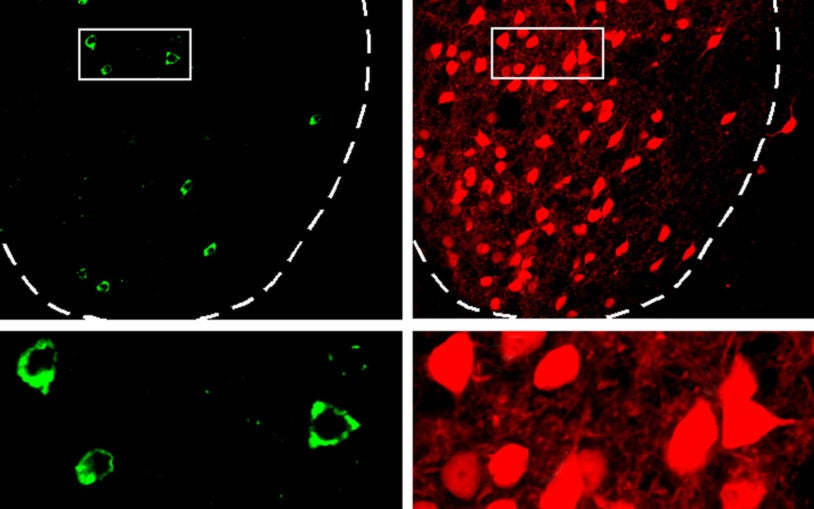
Neuronal circuits filter out distractions in the brain
December 15, 2014
Scientists identify a neural pathway that controls attention, with implications for psychiatric disorders
4 CSHL scientists will contribute to President’s BRAIN Initiative under new NIH grants
September 30, 2014
Four scientists at Cold Spring Harbor Laboratory (CSHL) will perform research in President Obama’s BRAIN Initiative under two new CSHL grants
A shift in the code: New method reveals hidden genetic landscape
August 18, 2014
Scientists develop algorithm to uncover genomic insertions and deletions involved in autism, OCD
Unprecedented detail of intact neuronal receptor offers blueprint for drug developers
May 29, 2014
NMDA receptor malfunction is implicated in Alzheimer’s, Parkinson’s, depression, schizophrenia, autism, and stroke
Dealing with stress—to cope or to quit?
May 27, 2014
Researchers identify neurons that determine whether an individual will be depressed or resilient
Research sees overlap in altered genes found in schizophrenia, autism and intellectual disability
April 28, 2014
New evidence supports theory that some cases of schizophrenia and autism have malfunctions in some of the same genes are contributing to pathology.
Scientists discover a new pathway for fear deep within the brain
February 12, 2014
‘Far-reaching’ neurons connect the amygdala with fear response center to control behavior
Team demonstrates power of precision medicine in successful treatment of patient with disabling OCD
October 3, 2013
Psychiatrist, geneticist, neurosurgeon & genome sequencers generate data on one patient, with powerful implications for care and counseling.
CSHL’s sequencing power gets a fresh boost
August 12, 2011
W. Richard McCombie discusses the evolution of genomic sequencing technology and the use of new third generation machines.
Collaboration column: SMA therapeutics and potential drug target for schizophrenia
March 11, 2011
CSHL's Adrian Krainer works on treatment for spinal muscular atrophy that reverses the disease's symptoms.
Cold Spring Harbor Laboratory surpasses capital campaign goal
July 15, 2009
Over $340 million raised for research and education
Researchers at CSHL began working on the genetics of cognitive disorders in 2005 following a generous gift from Ted and Vada Stanley. Under the leadership of James Watson, this effort grew substantially in 2007 with a gift from the Stanley’s to establish the Stanley Institute for Cognitive Genomics. At that time, new DNA sequencing technologies, called “Next Generation Sequencing” were being developed. CSHL Professor and sequencing pioneer W. Richard McCombie was one of the first to apply this technology to study the genetics of cognitive disorders putting the Stanley Institute on the map.
With the goal of improving diagnosis and the potential for personalized therapy, the Stanley Center began applying state-of-the-art genomics technologies to identify genetic variants contributing to schizophrenia, bipolar disorder and major depression. Together with collaborators in the U.S., Scotland, Ireland, Pakistan and Australia, CSHL focused first on sequencing the complete genomes or protein coding regions of the genomes of families that had many members suffering from mental illness. One of the key findings demonstrated an overlap among genes contributing to schizophrenia and autism. CSHL also showed a link between the genes involved in the modification of chromatin structure and schizophrenia.
In 2014, with continuing support from Ted and Vada Stanley, the Stanley Institute incorporated the efforts of CSHL neuroscientists focused on understanding how brain circuits assemble and function. Like the genetics group, neuroscientists are developing and applying new technologies, including sophisticated brain mapping technologies and novel behavioral paradigms for studying decision making, attention and other processes. Stanley Institute neuroscientists are now studying the function of genes that have been implicated in cognitive disorders.
Roberts, N. J., Norris, A. L., Petersen, G. M., Bondy, M. L., Brand, R., Gallinger, S., Kurtz, R. C., Olson, S. H., Rustgi, A. K., Schwartz, A. G., Stoffel, E. M., Syngal, S., Zogopoulos, G., Ali, S. Z., Axilbund, J., Chaffee, K. G., Chen, Y. C., Cote, M. L., Childs, E. J., Douville, C., Goes, F. S., Herman, J. M., Iacobuzio-Donahue, C., Kramer, M., Makohon-Moore, A., McCombie, R. W., McMahon, K. W., Niknafs, N., Parla, J., Pirooznia, M., Potash, J. B., Rhim, A. D., Smith, A. L., Wang, Y., Wolfgang, C. L., Wood, L. D., Zandi, P. P., Goggins, M., Karchin, R., Eshleman, J. R., Papadopoulos, N., Kinzler, K. W., Vogelstein, B., Hruban, R. H. , Klein, A. P. (2016) Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov, 6(2) pp. 166-175.
Jiménez-Barrón, Laura T., O'Rawe, Jason A., Wu, Yiyang, Yoon, Margaret, Fang, Han, Iossifov, Ivan , Lyon, Gholson J. (2015) Genome-wide variant analysis of simplex autism families with an integrative clinical-bioinformatics pipeline. Molecular Case Studies, 1(1)
Guo, Y., Ding, X., Shen, Y., Lyon, G. J. , Wang, K. (2015) SeqMule: automated pipeline for analysis of human exome/genome sequencing data. Sci Rep, 5 pp. 14283.
Regan, Michael C., Romero-Hernandez, Annabel , Furukawa, Hiro (2015) A structural biology perspective on NMDA receptor pharmacology and function. Current Opinion in Structural Biology, 33 pp. 68-75.
Doerfel, Max , Lyon, Gholson J. (2015) The biological functions of Naa10 – from amino-terminal acetylation to human disease. Gene, 567(2) pp. 103-131.
Krishnan, K., Wang, B. S., Lu, J., Wang, L., Maffei, A., Cang, J. , Huang, Z. J. (2015) MeCP2 regulates the timing of critical period plasticity that shapes functional connectivity in primary visual cortex. Proc Natl Acad Sci U S A, 112(34) pp. E4782-E4791.
Karakas, Erkan, Regan, Michael C. , Furukawa, Hiro (2015) Emerging structural insights into the function of ionotropic glutamate receptors. Trends in Biochemical Sciences, 40(6) pp. 328-337.
Lazarus, M. S., Krishnan, K. , Huang, Z. J. (2015) GAD67 Deficiency in Parvalbumin Interneurons Produces Deficits in Inhibitory Transmission and Network Disinhibition in Mouse Prefrontal Cortex. Cerebral Cortex, 25(5) pp. 1290-1296.
Delevich, K., Tucciarone, J., Huang, Z. J. , Li, B. (2015) The Mediodorsal Thalamus Drives Feedforward Inhibition in the Anterior Cingulate Cortex via Parvalbumin Interneurons. Journal of Neuroscience , 35(14) pp. 5743-53.
Hirschtritt, M. E., Lee, P. C., Pauls, D. L., Dion, Y., Grados, M. A., Illmann, C., King, R. A., Sandor, P., McMahon, W. M., Lyon, G. J., Cath, D. C., Kurlan, R., Robertson, M. M., Osiecki, L., Scharf, J. M. , Mathews, C. A. (2015) Lifetime Prevalence, Age of Risk, and Genetic Relationships of Comorbid Psychiatric Disorders in Tourette Syndrome. JAMA Psychiatry, 72(4) pp. 325.
Penzo, M. A., Robert, V., Tucciarone, J., De Bundel, D., Wang, M., Van Aelst, L., Darvas, M., Parada, L. F., Palmiter, R. D., He, M., Huang, Z. J. , Li, B. (2015) The paraventricular thalamus controls a central amygdala fear circuit. Nature, 519(7544) pp. 455-459.
Verleyen, W., Ballouz, S. , Gillis, J. (2015) Measuring the wisdom of the crowds in network-based gene function inference. Bioinformatics, 31(5) pp. 745-752.
Ballouz, S., Verleyen, W. , Gillis, J. (2015) Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics,
Perova, Z., Delevich, K. , Li, B. (2015) Depression of excitatory synapses onto parvalbumin interneurons in the medial prefrontal cortex in susceptibility to stress. Journal of Neuroscience, 35(7) pp. 3201-3206.
Ahrens, S., Jaramillo, S., Yu, K., Ghosh, S., Hwang, G., Paik, R., Lai, C., He, M., Huang, Z. J. , Li, B. (2015) ErbB4 regulation of a thalamic reticular nucleus circuit for sensory selection. Nature Neuroscience, 18 pp. 104-111.
He, M., Person, T. N., Hebbring, S. J., Heinzen, E., Ye, Z., Schrodi, S. J., McPherson, E. W., Lin, S. M., Peissig, P. L., Brilliant, M. H., O'Rawe, J., Robison, R. J., Lyon, G. J. , Wang, K. (2015) SeqHBase: a big data toolset for family based sequencing data analysis. Journal of Medical Genetics, 52(4) pp. 282-288.
O'Rawe, J. A., Ferson, S. , Lyon, G. J. (2015) Accounting for uncertainty in DNA sequencing data. Trends Genet, 31(2) pp. 61-68.
Kim, Yongsoo, Venkataraju, Kannan Umadevi, Pradhan, Kith, Mende, Carolin, Taranda, Julian, Turaga, Srinivas C, Arganda-Carreras, Ignacio, Ng, Lydia, Hawrylycz, Michael J, Rockland, Kathleen S, Seung, H. Sebastian , Osten, Pavel (2015) Mapping Social Behavior-Induced Brain Activation at Cellular Resolution in the Mouse. Cell Reports, 10(2) pp. 292-305.
O'Rawe, Jason, Wu, Yiyang, Rope, Alan, Jimenez Barron, Laura, Swensen, Jeffrey J., Fang, Han, Mittelman, David, Highnam, Gareth, Robison, Reid J., Yang, Edward, Wang, Kai , Lyon, Gholson (2015) A variant in TAF1 is associated with a new syndrome with severe intellectual disability and characteristic dysmorphic features. bioRxiv,
Myklebust, L. M., Van Damme, P., Stove, S. I., Dorfel, M. J., Abboud, A., Kalvik, T. V., Grauffel, C., Jonckheere, V., Wu, Y., Swensen, J., Kaasa, H., Liszczak, G., Marmorstein, R., Reuter, N., Lyon, G. J., Gevaert, K. , Arnesen, T. (2014) Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Human Molecular Genetics, 24(7) pp. 1956-1976.
Iossifov, I., O'Roak, B. J., Sanders, S. J., Ronemus, M., Krumm, N., Levy, D., Stessman, H. A., Witherspoon, K. T., Vives, L., Patterson, K. E., Smith, J. D., Paeper, B., Nickerson, D. A., Dea, J., Dong, S., Gonzalez, L. E., Mandell, J. D., Mane, S. M., Murtha, M. T., Sullivan, C. A., Walker, M. F., Waqar, Z., Wei, L., Willsey, A. J., Yamrom, B., Lee, Y. H., Grabowska, E., Dalkic, E., Wang, Z., Marks, S., Andrews, P., Leotta, A., Kendall, J., Hakker, I., Rosenbaum, J., Ma, B., Rodgers, L., Troge, J., Narzisi, G., Yoon, S., Schatz, M. C., Ye, K., McCombie, W. R., Shendure, J., Eichler, E. E., State, M. W. , Wigler, M. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature, 515(7526) pp. 216-221.
Pirooznia, M., Kramer, M., Parla, J., Goes, F. S., Potash, J. B., McCombie, W. R. , Zandi, P. P. (2014) Validation and assessment of variant calling pipelines for next-generation sequencing. Human Genomics, 8
McCarthy, S. E., Gillis, J., Kramer, M., Lihm, J., Yoon, S., Berstein, Y., Mistry, M., Pavlidis, P., Solomon, R., Ghiban, E., Antoniou, E., Kelleher, E., O'Brien, C., Donohoe, G., Gill, M., Morris, D. W., McCombie, W. R. , Corvin, A. (2014) De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Molecular Psychiatry, 19(6) pp. 652-658.
Thomson, P. A., Parla, J. S., McRae, A. F., Kramer, M., Ramakrishnan, K., Yao, J., Soares, D. C., McCarthy, S., Morris, S. W., Cardone, L., Cass, S., Ghiban, E., Hennah, W., Evans, K. L., Rebolini, D., Millar, J. K., Harris, S. E., Starr, J. M., MacIntyre, D. J., McIntosh, A. M., Watson, J. D., Deary, I. J., Visscher, P. M., Blackwood, D. H., McCombie, W. R. , Porteous, D. J. (2014) 708 Common and 2010 rare DISC1 locus variants identified in 1542 subjects: Analysis for association with psychiatric disorder and cognitive traits. Molecular Psychiatry, 19(6) pp. 668-675.
Karakas, E. , Furukawa, H. (2014) Crystal structure of a heterotetrameric NMDA receptor ion channel. Science, 344(6187) pp. 992-7.
Wang, M., Perova, Z., Arenkiel, B. R. , Li, B. (2014) Synaptic modifications in the medial prefrontal cortex in susceptibility and resilience to stress. Journal of Neuroscience, 34(22) pp. 7485-92.
Li, W., Wu, J., Kim, S. Y., Zhao, M., Hearn, S. A., Zhang, M. Q., Meistrich, M. L. , Mills, A. A. (2014) Chd5 orchestrates chromatin remodelling during sperm development. Nature Communications, 5 pp. 3812.
Penzo, M. A., Robert, V. , Li, B. (2014) Fear Conditioning Potentiates Synaptic Transmission onto Long-Range Projection Neurons in the Lateral Subdivision of Central Amygdala. Journal of Neuroscience, 34(7) pp. 2432-7.
Gillis, J., Ballouz, S. , Pavlidis, P. (2014) Bias tradeoffs in the creation and analysis of protein-protein interaction networks. Journal of Proteomics, 100 pp. 44-54.
Jespersen, A., Tajima, N., Fernandez-Cuervo, G., Garnier-Amblard, E. C. , Furukawa, H. (2014) Structural Insights into Competitive Antagonism in NMDA Receptors. Neuron, 81(2) pp. 366-78.
Yao, J., Zhang, K. X., Kramer, M., Pellegrini, M. , McCombie, W. R. (2014) FamAnn: an automated variant annotation pipeline to facilitate target discovery for family-based sequencing studies. Bioinformatics,
Fang, Han, Wu, Yiyang, Narzisi, G., O'Rawe, Jason, Jimenez Barron, Laura, Rosenbaum, J., Ronemus, M., Iossifov, I., Schatz, M. C. , Lyon, Gholson J. (2014) Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med, 6(10) pp. 89.
Paul, S., Kuo, A., Schalch, T., Vogel, H., Joshua-Tor, L., McCombie, W. R., Gozani, O., Hammell, M. , Mills, A. A. (2013) Chd5 Requires PHD-Mediated Histone 3 Binding for Tumor Suppression. Cell Reports, 3(1) pp. 92-102.
Mistry, M., Gillis, J. , Pavlidis, P. (2013) Genome-wide expression profiling of schizophrenia using a large combined cohort. Molecular Psychiatry, 18(2) pp. 215-225.
Ragan, T., Kadiri, L. R., Venkataraju, K. U., Bahlmann, K., Sutin, J., Taranda, J., Arganda-Carreras, I., Kim, Y., Seung, H. S. , Osten, P. (2012) Serial two-photon tomography for automated ex vivo mouse brain imaging. Nature Methods, 9(3) pp. 255-258.
Horev, G., Ellegood, J., Lerch, J. P., Son, Y. E., Muthuswamy, L., Vogel, H., Krieger, A. M., Buja, A., Henkelman, R. M., Wigler, M. H. , Mills, A. A. (2011) Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proceedings of the National Academy of Sciences of the United States of America, 108(41) pp. 17076-81.
Parla, J. S., Iossifov, I., Grabill, I., Spector, M. S., Kramer, M. , McCombie, W. R. (2011) A comparative analysis of exome capture. Genome Biology, 12(9) pp. R97.
Malhotra, D., McCarthy, S., Michaelson, J. J., Vacic, V., Burdick, K. E., Yoon, S., Cichon, S., Corvin, A., Gary, S., Gershon, E. S., Gill, M., Karayiorgou, M., Kelsoe, J. R., Krastoshevsky, O., Krause, V., Leibenluft, E., Levy, D. L., Makarov, V., Bhandari, A., Malhotra, A. K., McMahon, F. J., Nöthen, M. M., Potash, J. B., Rietschel, M., Schulze, T. G. , Sebat, J. (2011) High frequencies of de novo cnvs in bipolar disorder and schizophrenia. Neuron, 72(6) pp. 951-963.
Additional materials of the author atCSHL Institutional Repository
Results
No staff found.