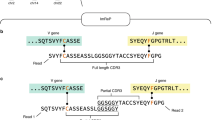
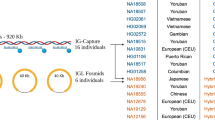
Abstract
Each B-cell receptor consists of a pair of heavy and light chains. High-throughput sequencing can identify large numbers of heavy- and light-chain variable regions (VH and VL) in a given B-cell repertoire, but information about endogenous pairing of heavy and light chains is lost after bulk lysis of B-cell populations. Here we describe a way to retain this pairing information. In our approach, single B cells (>5 × 104 capacity per experiment) are deposited in a high-density microwell plate (125 pl/well) and lysed in situ. mRNA is then captured on magnetic beads, reverse transcribed and amplified by emulsion VH:VL linkage PCR. The linked transcripts are analyzed by Illumina high-throughput sequencing. We validated the fidelity of VH:VL pairs identified by this approach and used the method to sequence the repertoire of three human cell subsets—peripheral blood IgG+ B cells, peripheral plasmablasts isolated after tetanus toxoid immunization and memory B cells isolated after seasonal influenza vaccination.
This is a preview of subscription content, access via your institution
Access options
Access to this article via Institution of Civil Engineers Library is not available.
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Reddy, S.T. et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat. Biotechnol. 28, 965–969 (2010).
Wu, X. et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011).
Ippolito, G.C. et al. Antibody repertoires in humanized NOD-scid-IL2R gamma(null) mice and human B cells reveals human-like diversification and tolerance checkpoints in the mouse. PLoS ONE 7, e35497 (2012).
Reddy, S.T. & Georgiou, G. Systems analysis of adaptive immunity by utilization of high-throughput technologies. Curr. Opin. Biotechnol. 22, 584–589 (2011).
Weinstein, J.A., Jiang, N., White, R.A., Fisher, D.S. & Quake, S.R. High-throughput sequencing of the zebrafish antibody repertoire. Science 324, 807–810 (2009).
Benichou, J., Ben-Hamo, R., Louzoun, Y. & Efroni, S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology 135, 183–191 (2012).
Fischer, N. Sequencing antibody repertoires: the next generation. MAbs 3, 17–20 (2011).
Wilson, P.C. & Andrews, S.F. Tools to therapeutically harness the human antibody response. Nat. Rev. Immunol. 12, 709–719 (2012).
Wardemann, H. et al. Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377 (2003).
Meijer, P. et al. Isolation of human antibody repertoires with preservation of the natural heavy and light chain pairing. J. Mol. Biol. 358, 764–772 (2006).
Smith, K. et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4, 372–384 (2009).
Frölich, D. et al. Secondary immunization generates clonally related antigen-specific plasma cells and memory B cells. J. Immunol. 185, 3103–3110 (2010).
Tanaka, Y. et al. Single-cell analysis of T-cell receptor repertoire of HTLV-1 tax-specific cytotoxic T cells in allogeneic transplant recipients with adult T-cell leukemia/lymphoma. Cancer Res. 70, 6181–6192 (2010).
Scheid, J.F. et al. Differential regulation of self-reactivity discriminates between IgG(+) human circulating memory B cells and bone marrow plasma cells. Proc. Natl. Acad. Sci. USA 108, 18044–18048 (2011).
Li, G.-M. et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc. Natl. Acad. Sci. USA 109, 9047–9052 (2012).
Sanchez-Freire, V., Ebert, A.D., Kalisky, T., Quake, S.R. & Wu, J.C. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat. Protoc. 7, 829–838 (2012).
White, A.K. et al. High-throughput microfluidic single-cell RT-qPCR. Proc. Natl. Acad. Sci. USA 108, 13999–14004 (2011).
Glanville, J. et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc. Natl. Acad. Sci. USA 108, 20066–20071 (2011).
Cheung, W.C. et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat. Biotechnol. 30, 447–452 (2012).
Sato, S. et al. Proteomics-directed cloning of circulating antiviral human monoclonal antibodies. Nat. Biotechnol. 30, 1039–1043 (2012).
Wrammert, J. et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671 (2008).
Scheid, J.F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011).
Seidl, K.J. et al. Frequent occurrence of identical heavy and light chain Ig rearrangements. Int. Immunol. 9, 689–702 (1997).
Ehrenmann, F., Kaas, Q. & Lefranc, M.P. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res. 38, D301–D307 (2010).
Lim, T.S. et al. V-gene amplification revisited. An optimised procedure for amplification of rearranged human antibody genes of different isotypes. N. Biotechnol. 27, 108–117 (2010).
Mei, H.E. et al. Blood-borne human plasma cells in steady state are derived from mucosal immune responses. Blood 113, 2461–2469 (2009).
Kyu, S.Y. et al. Frequencies of human influenza-specific antibody secreting cells or plasmablasts post vaccination from fresh and frozen peripheral blood mononuclear cells. J. Immunol. Methods 340, 42–47 (2009).
Mazor, Y., Barnea, I., Keydar, I. & Benhar, I. Antibody internalization studied using a novel IgG binding toxin fusion. J. Immunol. Methods 321, 41–59 (2007).
Friguet, B., Chaffotte, A.F., Djavadi-Ohaniance, L. & Goldberg, M.E. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods 77, 305–319 (1985).
Brochet, X., Lefranc, M.-P. & Giudicelli, V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–W508 (2008).
Acknowledgements
We thank B. Iverson and M. Pogson for insightful discussions and thoughtful advice, C. Das for assistance with antibody expression, and S. Reddy for help with initial studies. This work was funded by fellowships to B.J.D. from the Hertz Foundation, the University of Texas Donald D. Harrington Foundation and the National Science Foundation, and also by the US National Institutes of Health U19AI057234-09 (P.C.W.), U19AI057234-09 (G.G.), and U54AI057156 (G.G. and A.E.D.).
Author information
Authors and Affiliations
Contributions
B.J.D. and G.G. developed the methodology and designed the experiments. B.J.D., G.C.I. and G.G. wrote the manuscript; B.J.D., G.C.I., R.P.D., J.J.L., Y.W., B.M.R., C.G. and S.F.A. performed the experiments; B.J.D. carried out the bioinformatic analysis; S.P.H.-S. performed Illumina sequencing; G.C.I., N.V., T.D., P.C.W., C.G.W. and A.D.E. helped design experiments; B.J.D., G.C.I., J.J.L., Y.W., S.P.H.-S., A.D.E. and G.G. analyzed the data.
Corresponding author
Ethics declarations
Competing interests
G.G., B.J.D., A.D.E. and S.P.H.-S. declare competing financial interests in the form of a provisional patent application filed by the University of Texas, Austin.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–7 and Supplementary Figs. 1–3 (PDF 252 kb)
Supplementary Data
Supplementary Data sets (zip file) (ZIP 1607 kb)
Supplementary Video 1
Real-time video was used to observe MOPC-315 plasmacytoma cell lysis inside sealed microwells (MOV 9212 kb)
Rights and permissions
About this article
Cite this article
DeKosky, B., Ippolito, G., Deschner, R. et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol 31, 166–169 (2013). https://doi.org/10.1038/nbt.2492
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2492